A guide on regression error metrics (MSE, RMSE, MAE, MAPE, sMAPE, MPE) with Python code
2022-08-18
Setup collaborative MLflow with PostgreSQL as Tracking Server and MinIO as Artifact Store using docker containers
2022-08-30Pipeline, ColumnTransformer, and FeatureUnion are three powerful tools that anyone who wants to master using sklearn must know. It’s, therefore, crucial to learn how to use these efficiently when building a machine learning model.
Before we dive in, let’s first get aligned on two terms:
- Transformer: A transformer refers to an object with
fit()andtransform()method that cleans, reduces, expands, or generates features. Simply put, transformers help you transform your data towards the desired format for a machine learning model.OneHotEncoderandMinMaxScalerare examples of transformers. - Estimator: An estimator refers to a machine learning model. It is an object with
fit()andpredict()method. We will use estimator and model interchangeably throughout this post. Here are some example estimators.
Note: All of the codes in this post are available in a Jupyter Notebook on Github.
Prerequisites
If you want to follow along with the code on your computer, make sure you have pandas, seaborn, and sklearn installed. I have used and tested the scripts in Python 3.9 and scikit-learn==1.1.1 in Jupyter Notebook.
Let’s import the required packages and the dataset on restaurant tips. Details about this dataset including the data dictionary can be found here (this source is actually for R, but it appears to be referring to the same underlying dataset).
#Set seed
seed = 123
#Import package / module for data
import pandas as pd
from seaborn import load_dataset
#Importing modules for Feature Engineering and modeling
from sklearn.model_selection import train_test_split
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.preprocessing import OneHotEncoder, MinMaxScaler
from sklearn.impute import SimpleImputer
from sklearn.pipeline import Pipeline, FeatureUnion
from sklearn.compose import ColumnTransformer
from sklearn.linear_model import LinearRegression
#Loading data sets
df = load_dataset('tips').drop(columns=['tip', 'sex']).sample(n=5, random_state=seed)
#Add missing values
df.iloc[[1, 2, 4], [2, 4]] = np.nan
df
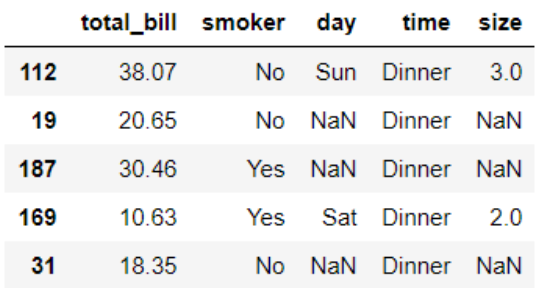
Using a handful of records will make it easy to monitor inputs and outputs at each step. Thus, we will only use a sample of 5 records from the dataset.
Scikit-Learn Pipeline
Let’s assume we wanted to use smoker, day, and time columns to predict total_bill. We will drop the size column and partition the data first:
#Partition data
X_train, X_test, y_train, y_test = train_test_split(df.drop(columns=['total_bill', 'size']),
df['total_bill'],
test_size=.2,
random_state=seed)
Typically, the raw data is not in a state where we can straight away feed it into a machine learning model. Therefore, transforming the data to a state that is acceptable and useful for a model becomes an essential prerequisite for modeling. Let’s do the following transformations as preparation:
- Impute missing values with ‘missing’
- One-hot encode them
Here’s one approach to complete these two steps:
#Input training data
imputer = SimpleImputer(strategy='constant', fill_value='missing')
X_train_imputed = imputer.fit_transform(X_train)
#Coding training data
encoder = OneHotEncoder(handle_unknown='ignore', sparse=False)
X_train_encoded = encoder.fit_transform(X_train_imputed)
#Check the data before and after the training
print("******************** Training data ********************")
display(X_train)
display(pd.DataFrame(X_train_imputed, columns=X_train.columns))
display(pd.DataFrame(X_train_encoded, columns=encoder.get_feature_names_out(X_train.columns)))
#Convert test data
X_test_imputed = imputer.transform(X_test)
X_test_encoded = encoder.transform(X_test_imputed)
#Check the data before and after the test
print("******************** Test data ********************")
display(X_test)
display(pd.DataFrame(X_test_imputed, columns=X_train.columns))
display(pd.DataFrame(X_test_encoded, columns=encoder.get_feature_names_out(X_train.columns)))
You may have noticed that we used column names from the training dataset when mapping back the column name for the test dataset. That’s because I prefer to use column names from the data that the transformers were trained on. However, it will give the same results if we use the test dataset.
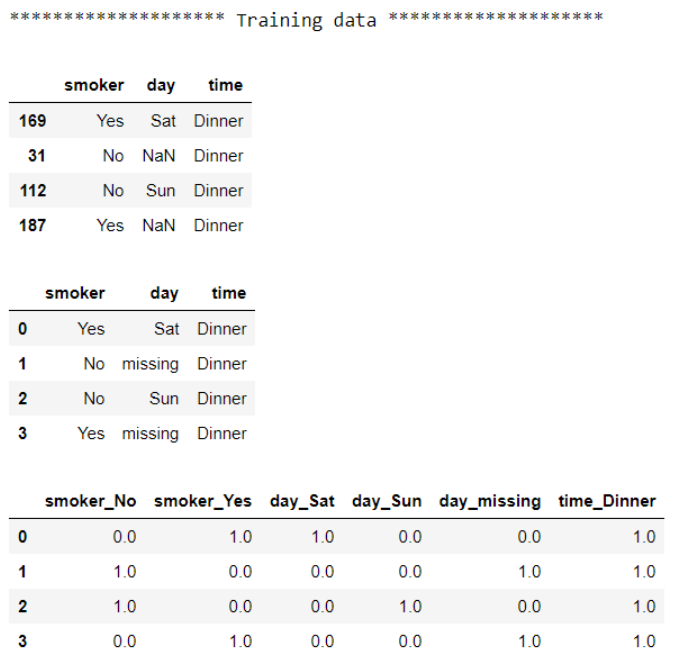
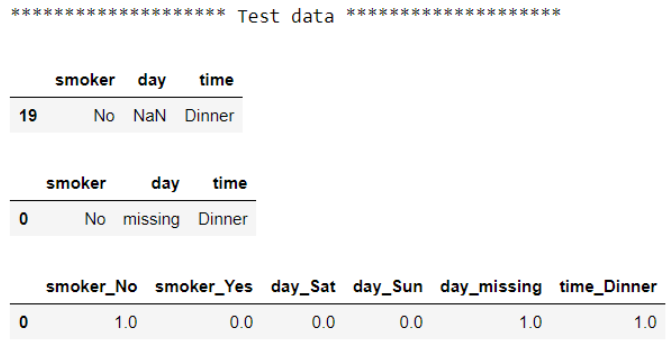
For each dataset, we see raw data first, then output after imputation, and the output after encoding last. This approach gets the job done. However, we manually feed output from the previous step to the next step as an input and have multiple interim outputs. We also had to repeat each step on the test data. As the number of steps increases, it will become more tedious to maintain and more prone to error. We can write more streamlined and concise code with Pipeline:
#Match pipes to training data
pipe = Pipeline([('imputer', SimpleImputer(strategy='constant', fill_value='missing')),
('encoder', OneHotEncoder(handle_unknown='ignore', sparse=False))])
pipe.fit(X_train)
#Check the data before and after the training
print("******************** Training data ********************")
display(X_train)
display(pd.DataFrame(pipe.transform(X_train), columns=pipe['encoder'].get_feature_names_out(X_train.columns)))
#Check the data before and after the test
print("******************** Test data ********************")
display(X_test)
display(pd.DataFrame(pipe.transform(X_test), columns=pipe['encoder'].get_feature_names_out(X_train.columns)))
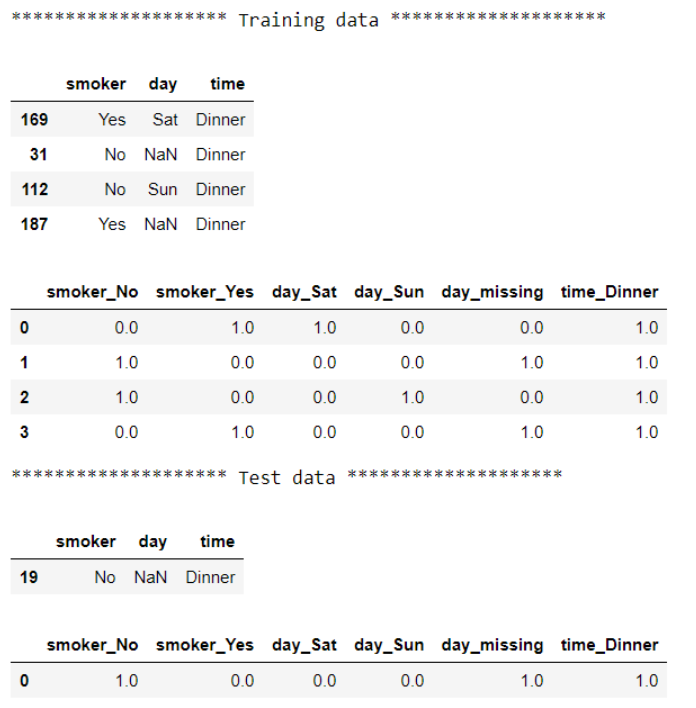
When using Pipeline, each step passes its output to the next step as an input. Therefore, we don’t have to manually keep track of different versions of the data. This approach gives us the exact same final output but with more elegant code.
Having looked at the transformed data, it’s time to add a model to our example. Let’s tweak the code to add a simple model to the first approach:
#Input training data
imputer = SimpleImputer(strategy='constant', fill_value='missing')
X_train_imputed = imputer.fit_transform(X_train)
#Coding training data
encoder = OneHotEncoder(handle_unknown='ignore', sparse=False)
X_train_encoded = encoder.fit_transform(X_train_imputed)
#Make the model fit the training data
model = LinearRegression()
model.fit(X_train_encoded, y_train)
#Forecast training data
y_train_pred = model.predict(X_train_encoded)
print(f"Predictions on training data: {y_train_pred}")
#Convert test data
X_test_imputed = imputer.transform(X_test)
X_test_encoded = encoder.transform(X_test_imputed)
#Forecast test data
y_test_pred = model.predict(X_test_encoded)
print(f"Predictions on test data: {y_test_pred}")

We will do the same for the approach with pipeline:
#Match pipes to training data
pipe = Pipeline([('imputer', SimpleImputer(strategy='constant', fill_value='missing')),
('encoder', OneHotEncoder(handle_unknown='ignore', sparse=False)),
('model', LinearRegression())])
pipe.fit(X_train, y_train)
#Forecast training data
y_train_pred = pipe.predict(X_train)
print(f"Predictions on training data: {y_train_pred}")
#Forecast test data
y_test_pred = pipe.predict(X_test)
print(f"Predictions on test data: {y_test_pred}")

You may have noticed how simple it is to make predictions once we train a pipeline. Calling a single line: pipe.predict(X) on raw data will do the transformations and then return a prediction. It’s also easy to see the sequence of steps too. Let’s summarise the two approaches visually:

Using Pipeline not only organizes and streamlines your code but also has many other benefits, here are some of them:
- Ability to fine-tune pipeline: When building a model, have you ever had to go back a step to try a different way to preprocess the data and run the model again to see if a tweak in preprocessing step improves the fitness of the model? When optimizing a model, the cogs are not only in the model hyperparameters but also in the implementation of preprocessing steps. With this in mind, when we have a single pipeline object that unifies transformers and an estimator, we can fine-tune the hyperparameters of the entire pipeline including both transformers and an estimator with either GridSearchCV or RandomizedSearchCV.
- Easier deployment: All transforming steps used to prepare the data when training a model should also be applied to the data in the production environment when making predictions. When we train a
Pipeline, we train a single object which contains data transformers and a model. Once trained, thisPipelineobject can be used for smoother deployment.
Scikit-Learn ColumnTransformer
In the previous example, we imputed and encoded all columns the same way. However, we often need to apply different sets of transformers to different groups of columns. For instance, we would want to apply OneHotEncoder to only categorical columns but not to numerical columns. This is where ColumnTransformer comes in. This time, we will partition the dataset keeping all columns so that we have both numerical and categorical features.
#Partition data
X_train, X_test, y_train, y_test = train_test_split(df.drop(columns=['total_bill']),
df['total_bill'],
test_size=.2,
random_state=seed)
#Define classification columns
categorical = list(X_train.select_dtypes('category').columns)
print(f"Categorical columns are: {categorical}")
#Define numeric columns
numerical = list(X_train.select_dtypes('number').columns)
print(f"Numerical columns are: {numerical}")
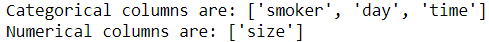
We have split the features into two groups based on the data type. Column groupings can be done based on what’s appropriate for the data. For instance, categorical columns could further be split into multiple groups if different preprocessing pipelines are more suitable for them.
The code from the previous section will no longer work now because we have multiple data types. Let’s see an example where we use ColumnTransformer together with Pipeline to do the same transformations as before in the presence of multiple data types.
#Define classification pipeline
cat_pipe = Pipeline([('imputer', SimpleImputer(strategy='constant', fill_value='missing')),
('encoder', OneHotEncoder(handle_unknown='ignore', sparse=False))])
#Make columntransformer fit training data
preprocessor = ColumnTransformer(transformers=[('cat', cat_pipe, categorical)],
remainder='passthrough')
preprocessor.fit(X_train)
#Ready to list
cat_columns = preprocessor.named_transformers_['cat']['encoder'].get_feature_names_out(categorical)
columns = np.append(cat_columns, numerical)
#Check the data before and after the training
print("******************** Training data ********************")
display(X_train)
display(pd.DataFrame(preprocessor.transform(X_train), columns=columns))
#Check the data before and after the test
print("******************** Test data ********************")
display(X_test)
display(pd.DataFrame(preprocessor.transform(X_test), columns=columns))
The output for categorical columns is identical to that of the previous section. The only difference is that this version has an additional column: size. We have passed cat_pipe (previously called pipe in section 1) to the ColumnTransformer to transform the categorical columns and specified remainder='passthrough' to keep the remaining column as it is.
Wouldn’t it be nice to also transform the numerical column too? In particular, let’s impute missing values with median size and scale it between 0 and 1:
#Define classification pipeline
cat_pipe = Pipeline([('imputer', SimpleImputer(strategy='constant', fill_value='missing')),
('encoder', OneHotEncoder(handle_unknown='ignore', sparse=False))])
#Define value pipeline
num_pipe = Pipeline([('imputer', SimpleImputer(strategy='median')),
('scaler', MinMaxScaler())])
#Make columntransformer fit training data
preprocessor = ColumnTransformer(transformers=[('cat', cat_pipe, categorical),
('num', num_pipe, numerical)])
preprocessor.fit(X_train)
#Ready to list
cat_columns = preprocessor.named_transformers_['cat']['encoder'].get_feature_names_out(categorical)
columns = np.append(cat_columns, numerical)
#Check the data before and after the training
print("******************** Training data ********************")
display(X_train)
display(pd.DataFrame(preprocessor.transform(X_train), columns=columns))
#Check the data before and after the test
print("******************** Test data ********************")
display(X_test)
display(pd.DataFrame(preprocessor.transform(X_test), columns=columns))
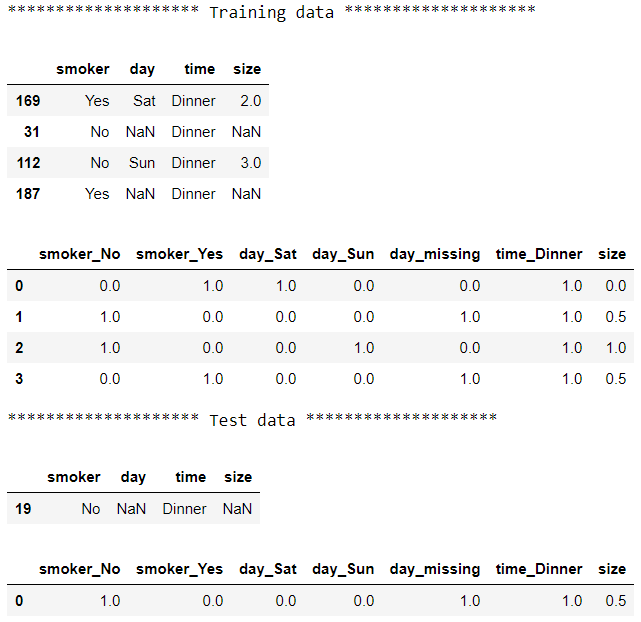
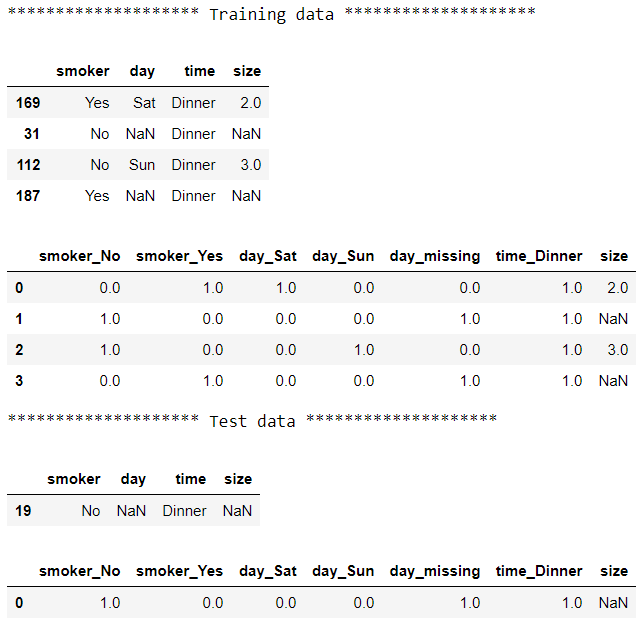
Now all the columns are imputed and range between 0 and 1. Using ColumnTransformer and Pipeline, we split the data into two groups, applied a different pipeline with different sets of transformers to each group then pasted the results together:
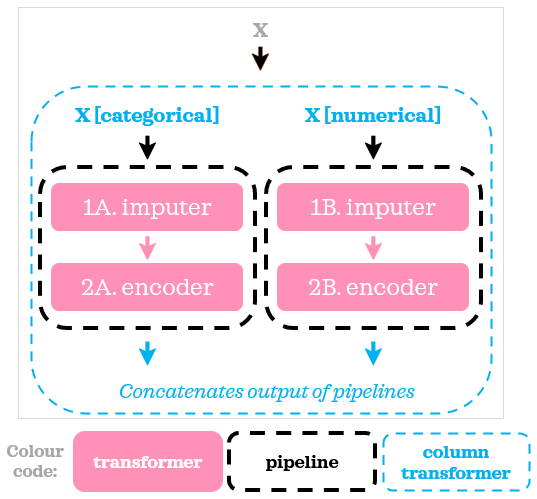
Even though we have the same number of steps in both numerical and categorical pipelines in our example, you can have any number of steps in the pipelines as they are independent of each other. Let’s now add a model to our example:
#Define classification pipeline
cat_pipe = Pipeline([('imputer', SimpleImputer(strategy='constant', fill_value='missing')),
('encoder', OneHotEncoder(handle_unknown='ignore', sparse=False))])
#Define value pipeline
num_pipe = Pipeline([('imputer', SimpleImputer(strategy='median')),
('scaler', MinMaxScaler())])
#Combined classification pipeline and numerical pipeline
preprocessor = ColumnTransformer(transformers=[('cat', cat_pipe, categorical),
('num', num_pipe, numerical)])
#Install transformer and training data estimator on the pipeline
pipe = Pipeline(steps=[('preprocessor', preprocessor),
('model', LinearRegression())])
pipe.fit(X_train, y_train)
#Forecast training data
y_train_pred = pipe.predict(X_train)
print(f"Predictions on training data: {y_train_pred}")
#Forecast test data
y_test_pred = pipe.predict(X_test)
print(f"Predictions on test data: {y_test_pred}")

To put together the preprocessing steps specified in ColumnTransformer with a model, we used a Pipeline on the outside. Here is its visual representation:

ColumnTransformer complements Pipeline nicely when we need to do different sets of operations on different subsets of columns.
Scikit-Learn FeatureUnion
Outputs following the code are omitted in this section because they are identical to that of section: 2. ColumnTransformer.
FeatureUnion is another useful tool. It is capable of doing what ColumnTransformer just did but in a longer way:
#Custom pipe
class ColumnSelector(BaseEstimator, TransformerMixin):
"""Select only specified columns."""
def __init__(self, columns):
self.columns = columns
def fit(self, X, y=None):
return self
def transform(self, X):
return X[self.columns]
#Define classification pipeline
cat_pipe = Pipeline([('selector', ColumnSelector(categorical)),
('imputer', SimpleImputer(strategy='constant', fill_value='missing')),
('encoder', OneHotEncoder(handle_unknown='ignore', sparse=False))])
#Define value pipeline
num_pipe = Pipeline([('selector', ColumnSelector(numerical)),
('imputer', SimpleImputer(strategy='median')),
('scaler', MinMaxScaler())])
#Featureunion fitting training data
preprocessor = FeatureUnion(transformer_list=[('cat', cat_pipe),
('num', num_pipe)])
preprocessor.fit(X_train)
#Ready to list
cat_columns = preprocessor.transformer_list[0][1][2].get_feature_names_out(categorical)
columns = np.append(cat_columns, numerical)
#Check the data before and after the training
print("******************** Training data ********************")
display(X_train)
display(pd.DataFrame(preprocessor.transform(X_train), columns=columns))
#Check the data before and after the test
print("******************** Test data ********************")
display(X_test)
display(pd.DataFrame(preprocessor.transform(X_test), columns=columns))
We can think of FeatureUnion as if it creates a copy of the data, transforms those copies in parallel, and then pastes together the results. The term copy here is more of an analogy to aid conceptualization than a technical reference.
At the beginning of each pipeline, we added an extra step where we selected relevant columns using a custom transformer: ColumnSelector in lines 14 and 19. Here’s how we can visually summarise the script above:
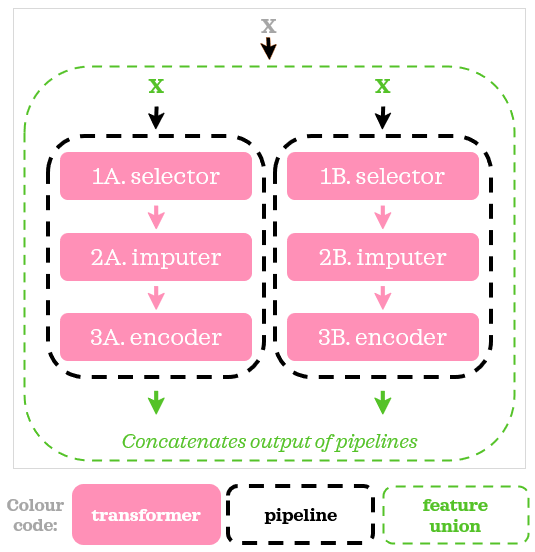
Now, it’s time to add a model to the script:
#Define classification pipeline
cat_pipe = Pipeline([('selector', ColumnSelector(categorical)),
('imputer', SimpleImputer(strategy='constant', fill_value='missing')),
('encoder', OneHotEncoder(handle_unknown='ignore', sparse=False))])
#Define value pipeline
num_pipe = Pipeline([('selector', ColumnSelector(numerical)),
('imputer', SimpleImputer(strategy='median')),
('scaler', MinMaxScaler())])
#Combined classification pipeline and numerical pipeline
preprocessor = FeatureUnion(transformer_list=[('cat', cat_pipe),
('num', num_pipe)])
#Combined classification pipeline and numerical pipeline
pipe = Pipeline(steps=[('preprocessor', preprocessor),
('model', LinearRegression())])
pipe.fit(X_train, y_train)
#Forecast training data
y_train_pred = pipe.predict(X_train)
print(f"Predictions on training data: {y_train_pred}")
#Forecast test data
y_test_pred = pipe.predict(X_test)
print(f"Predictions on test data: {y_test_pred}")
It looks very similar to what we did with ColumnTransformer.
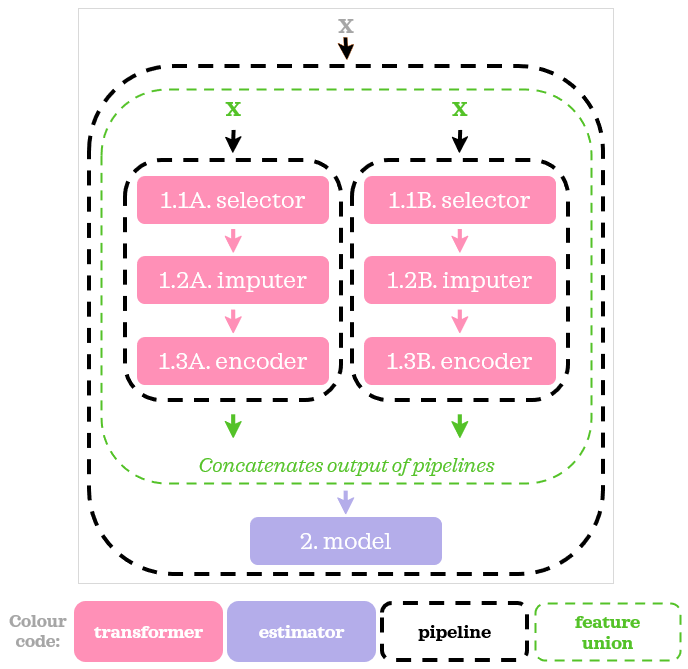
Difference Between ColumnTransformerFeatureUnion
ColumnTransformerBoth of these methods are used to combine independent transformations (transformers) into a single transformer, by independent I mean transformations (transformers) that don’t need to be executed sequentially, they will be executed in parallel and the output of each transformation will be merged at the end.
The main difference is that each transformer in a feature union object gets the whole dataset as input. While in the column transformer object, they get only part of the data as input. FeatureUnion applies different transformers to the whole of the input data and then combines the results by concatenating them. ColumnTransformer, on the other hand, applies different transformers to different subsets of the whole input data and again concatenates the results.
As seen in the previous example, using FeatureUnion is more verbose than using ColumnTransformer. Therefore, in my opinion, it’s better to use ColumnTransformer in a case similar to this. However, FeatureUnion definitely has its place. If you ever need to transform the same input data in different ways and use them as features, FeatureUnion is the one. For example, if you are working on text data and want to do both tf-idf vectorization of the data as well as extract the length of text, FeatureUnion is the perfect tool.
Bonus Example: Build NLP pipeline
Let’s imagine you have preprocessed your text data into a matrix using a vectorizer (e.g. TfidfVectorizer, CountVectorizer) as a preparation for a model. You have an inkling that deriving other features like the length of a document could also be useful to your model. While there are many possible ways to combine these preprocessing steps, leveraging FeatureUnion, ColumnTransformer, and/or Pipeline is probably one of the best approaches to accomplish the task. We will look at a few examples of how we can apply these awesome tools (with the main focus on FeatureUnion) in this section.
While it’s not necessary, having some understanding of the following topics may be helpful while/before reading this article.
◻️ TfidfVectorizer
◻️ Pipeline, ColumnTransformer and FeatureUnion
Import data
Let’s start by importing the necessary libraries and creating a minimal workable toy dataset on restaurant reviews:
import numpy as np
import pandas as pd
from nltk.tokenize import RegexpTokenizer
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import MinMaxScaler
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.compose import ColumnTransformer
from sklearn.pipeline import Pipeline, FeatureUnion
# Create a small sample data
X_train = pd.DataFrame(data={'document': ['Food was good.',
'Superb food :)',
'Absolutely superb!']})
X_train

Keeping the dataset small will help understand the process better and focus more on the practical applications of the actual sklearn tools.
Parallel transformers
a) Without Pipeline
Let’s imagine our goal for preprocessing is to vectorize the text and extract two features from the text. To be more precise, we want to do the following:
◻️ ️Vectorise the ‘document’ column with TfidfVectorizer◻️️ Add a feature showing the length of a text with CharacterCounter
◻️️ Add a feature showing the number of alphanumeric tokens in a text with TokenCounter
CharacterCounter and TokenCounter are custom transformers we are going to create. We can’t chain these three sequentially in a single Pipeline because TfidfVectorizer takes only the text column as an input and outputs a sparse matrix without the original text column. FeatureUnion is the perfect tool in this kind of situation. As FeatureUnion acts as if it creates copies of the input data, we can design the preprocessing flow like this with parallel transformers:

Each stream takes the same input: X[‘document’] and output is concatenated for us. For brevity and clarity, let’s refer to these streams as stream A, stream B, and stream C from here onwards. Now, let’s translate the flow into a code:
# Define custom transformers
class CharacterCounter(BaseEstimator, TransformerMixin):
"""Count the number of characters in a document."""
def __init__(self):
pass
def fit(self, X, y=None):
return self
def transform(self, X):
n_characters = X.str.len()
return n_characters.values.reshape(-1, 1) # 2D array
class TokenCounter(BaseEstimator, TransformerMixin):
"""Count the number of tokens in a document."""
def __init__(self):
pass
def fit(self, X, y=None):
self.tokeniser = RegexpTokenizer(r'[A-Za-z]+')
return self
def transform(self, X):
n_tokens = X.apply(lambda document: len(
self.tokeniser.tokenize(document)))
return n_tokens.values.reshape(-1, 1) # 2D array
# Build a FeatureUnion
text = 'document'
vectoriser = TfidfVectorizer(token_pattern=r'[a-z]+', stop_words='english')
character_counter = CharacterCounter()
token_counter = TokenCounter()
preprocessor = FeatureUnion([
('vectoriser', vectoriser),
('character_counter', character_counter),
('token_counter', token_counter)
])
preprocessor.fit(X_train[text])
# Transform the data and format for readibility
columns = np.append(preprocessor.transformer_list[0][1].get_feature_names_out(), [
'n_characters', 'n_tokens'])
X_train_transformed = pd.DataFrame(preprocessor.transform(X_train[text]).toarray(),
columns=columns)
X_train_transformed
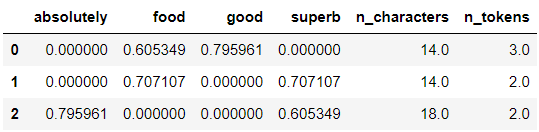
Tada! We have preprocessed the data using FeatureUnion.
b) With Pipeline
Let’s extend our previous example with Pipeline. This time, we will extend stream B and stream C with scalers. In other words, below summarises our goal:
◻️️ Vectorise the ‘document’ column with TfidfVectorizer◻️️ Add a feature showing the length of a text with CharacterCounter, then scale with MinMaxScaler
◻️️ Add a feature showing the number of alphanumeric tokens in a text with TokenCounter, then scale with MinMaxScaler
Using Pipeline, the preprocessing flow now can be summarised as follows:
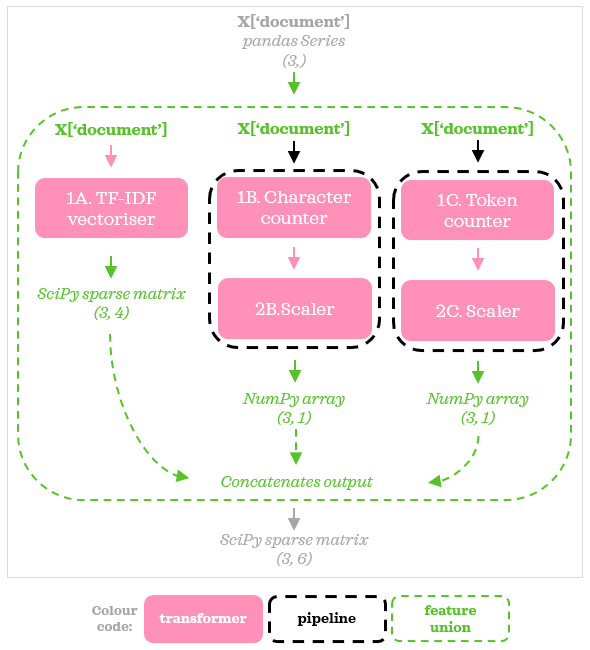
Pipelines give us the flexibility to extend any stream with more transformers. While in this example, both streams B & C used the same scaler, they don’t have to be the same since they are independent of each other. So now the code changes to:
# Build a FeatureUnion with Pipelines
character_pipe = Pipeline([
('character_counter', CharacterCounter()),
('scaler', MinMaxScaler())
])
token_pipe = Pipeline([
('token_counter', TokenCounter()),
('scaler', MinMaxScaler())
])
preprocessor = FeatureUnion([
('vectoriser', vectoriser),
('character', character_pipe),
('token', token_pipe)
])
preprocessor.fit(X_train[text])
# Transform the data and format for readibility
columns = np.append(preprocessor.transformer_list[0][1].get_feature_names_out(
), ['n_characters', 'n_tokens'])
X_train_transformed = pd.DataFrame(preprocessor.transform(X_train[text]).toarray(),
columns=columns)
X_train_transformed
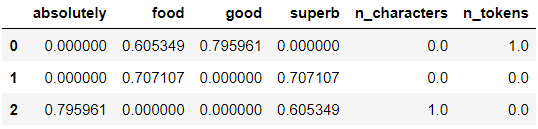
As we can see, FeatureUnion helps us keep the preprocessing steps organized and write clean code.
Sequential transformers
Now let’s look at another way to get the same output from previous snippets. Instead of adding parallel transformers, we can combine stream B and stream C and position them sequentially like this in a Pipeline:
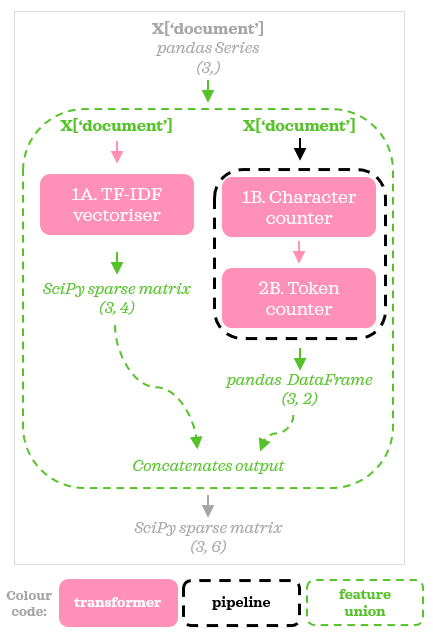
For this flow to work, we need to tweak our custom transformers and create a pipeline for them called counter_pipe and update the preprocessor:
# Define sample customer transformers
class CharacterCounter(BaseEstimator, TransformerMixin):
"""Count the number of characters in a document."""
def __init__(self):
pass
def fit(self, X, y=None):
return self
def transform(self, X):
n_characters = X.str.len().rename('n_characters')
df = pd.concat([X, n_characters], axis=1)
return df
class TokenCounter(BaseEstimator, TransformerMixin):
"""Count the number of tokens in a document."""
def __init__(self, col=None):
self.col = col
def fit(self, X, y=None):
self.tokeniser = RegexpTokenizer(r'[A-Za-z]+')
return self
def transform(self, X):
df = X.copy()
df['n_tokens'] = df[self.col].apply(lambda document: len(self.tokeniser.tokenize(document)))
return df.drop(columns=self.col)
# Build a FeatureUnion with Pipeline
counter_pipe = Pipeline([
('character_counter', CharacterCounter()),
('token_counter', TokenCounter(text))
])
preprocessor = FeatureUnion([
('vectoriser', vectoriser),
('counter', counter_pipe)
])
preprocessor.fit(X_train[text])
# Transform the data and format for readibility
columns = np.append(preprocessor.transformer_list[0][1].get_feature_names_out(
), ['n_characters', 'n_tokens'])
X_train_transformed = pd.DataFrame(preprocessor.transform(X_train[text]).toarray(),
columns=columns)
X_train_transformed
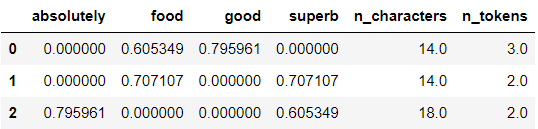
We can design the preprocessing flow in different ways while still getting the same results. In this particular example, I prefer the previous approach compared to this alternative approach. In general, it comes down to the use case and personal preference.
With ColumnTransformer
Lastly, this example is my favorite and the most exciting one out of all the examples we cover in this post. We will leverage all three tools: FeatureUnion, ColumnTransformer and Pipeline where each one plays a unique role and complements each other.
In practice, it’s not uncommon that the data to contain both text and non-text columns. For instance, text reviews may also be accompanied by star reviews. In this kind of scenario, we will look at how we can preprocess all so that we don’t discard potentially valuable information. Let’s expand the dataset with a few numerical features.
# Add two numerical columns
X_train['has_tipped'] = [1, 0, 1]
X_train['rating'] = [4, np.nan, 5]
X_train

Now our goal for preprocessing is to:
◻️️ Vectorise ‘document’ column with TfidfVectorizer◻️️ Add a feature showing the length of a text with CharacterCounter
◻️️ Add a feature showing the number of alphanumeric tokens in a text with TokenCounter
◻️️ Impute ‘has_tipped’ and ‘rating’ columns with median using SimpleImputer, then scale them with MinMaxScaler
So now the preprocessing flow can be visualized as follows:

Using ColumnTransformer, we split the dataset into two streams. The left stream has a Pipeline with two transformers. The right stream is identical to the FeatureUnion we looked at in the previous examples. Using the transformers we defined in the previous section, we can define the new flow like this:
# Build a ColumnTransformer with FeatureUnion
numerical = ['has_tipped', 'rating']
text_union = FeatureUnion([
('vectoriser', vectoriser),
('counter', counter_pipe)
])
num_pipe = Pipeline([
('imputer', SimpleImputer(strategy='median')),
('scaler', MinMaxScaler())
])
preprocessor = ColumnTransformer([
('num', num_pipe, numerical),
('text', text_union, text)
])
preprocessor.fit(X_train)
# Transform the data and format for readibility
terms = preprocessor.named_transformers_[
'text'].transformer_list[0][1].get_feature_names_out()
columns = np.concatenate((numerical, terms, ['n_characters', 'n_tokens']))
X_train_transformed = pd.DataFrame(
preprocessor.transform(X_train), columns=columns)
X_train_transformed

Voila! Hopefully, this shows a practical use case of how FeatureUnion, ColumnTransformer, Pipeline can work harmoniously with each other.
Final remarks on NLP pipeline examples
Applying FeatureUnion, ColumnTransformer, and Pipeline skillfully is a useful skill for Data Scientists. With these tools, the preprocessing process can be kept neat and organized. For instance, preprocessing unseen data is a breeze for all examples covered and can be accomplished in a single line: preprocessor.transform(new_data). Below shows an example for section 3:
# Create sample test data
X_test = pd.DataFrame({"document": ["Absolutely fantastic food"],
"has_tipped": [1],
"rating": [np.nan]})
# Preprocess the test data
preprocessor.transform(X_test)

Summary
As you may have noticed, Pipeline is the superstar. ColumnTransformer and FeatureUnion are additional tools to use with Pipeline. ColumnTransformer is more suitable when we want to divide and conquer in parallel whereas FeatureUnion allows us to apply multiple transformers on the same input data in parallel.
Here’s a simplified summary:

Resources:
https://towardsdatascience.com/pipeline-columntransformer-and-featureunion-explained-f5491f815f
https://medium.com/vickdata/a-simple-guide-to-scikit-learn-pipelines-4ac0d974bdcf
More examples at:
https://towardsdatascience.com/getting-the-most-out-of-scikit-learn-pipelines-c2afc4410f1a
https://medium.com/bigdatarepublic/integrating-pandas-and-scikit-learn-with-pipelines-f70eb6183696
https://www.kaggle.com/code/baghern/a-deep-dive-into-sklearn-pipelines/notebook
https://queirozf.com/entries/scikit-learn-pipeline-examples
http://zacstewart.com/2014/08/05/pipelines-of-featureunions-of-pipelines.html
https://michelleful.github.io/code-blog/2015/06/20/pipelines/
https://towardsdatascience.com/step-by-step-tutorial-of-sci-kit-learn-pipeline-62402d5629b6